Dual Index Sequencing
Accurate Demultiplexing
Unique dual indexes reduce per-sample cost and sequence libraries with the highest accuracy compared to other indexing strategies

Understanding Dual Index Sequencing
Unique dual indexes allow you to increase the number of samples sequenced per run and reduce per-sample cost compared to other indexing strategies. With just one unique dual index plate, you can pool 96 samples together.
Dual Index Sequencing Strategies
In addition to unique dual indexes, another strategy for dual index sequencing is to use combinatorial dual indexes. When preparing libraries for multiplexing, we recommend using unique dual indexes. With a dual-indexed library, the sequencing run includes two additional reads. The additional reads allow the post-run analysis software to demultiplex with increased accuracy.
Unique Dual Indexes
To prevent repeated sequences in a well plate, unique dual indexing uses unique identifiers on both ends of the sample.
One unique dual index plate enables 96 samples to be pooled together using 96 unique index 1 (i7) adapters and 96 unique index 2 (i5) adapters.
Combinatorial Dual Indexes
With combinatorial dual indexes, sequences are repeated across the rows and columns of a well plate.
Indexing is limited to index combinations of 8 unique dual pairs, so the index adapters share some sequences. Most libraries share sequences on the i7 or i5 end.
Why Use Unique Dual Indexes?
The benefits of unique dual indexing include:
- Greater efficiency for multiplexing
- Reduced per-sample cost by allowing more indexes to be included in a sequencing run compared to combinatorial dual or single indexes
- Highly purified and manufactured under Good Manufacturing Practice (GMP) conditions
- Improved design with color balance in mind
- Mitigates index hopping by filtering hopped reads
Learn more about how unique dual indexes can help ensure high accuracy on VR���˲�Ʊ sequencing systems.
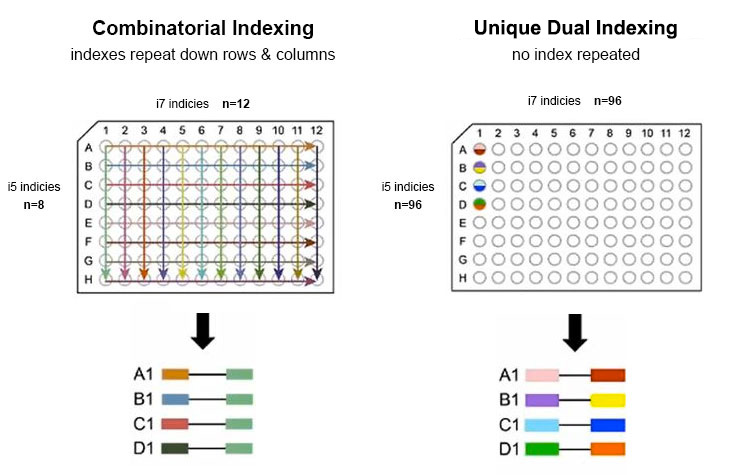
Comparison of Unique Dual and Combinatorial Dual Index Adapters
One unique dual index plate enables 96 samples to be pooled together. In contrast, combinatorial dual indexing is limited to index combinations of 8 unique dual pairs.
Recommended Index Adapters and Library Prep Kits for Key Applications
| Application | Library Prep Kits | Recommended Indexing Solutions |
|---|---|---|
| Whole-genome sequencing | VR���˲�Ʊ DNP Prep | IDT for VR���˲�Ʊ UD Indexes |
| RNA sequencing | VR���˲�Ʊ Stranded mRNA Prep VR���˲�Ʊ Stranded Total RNA Prep |
IDT for VR���˲�Ʊ – RNA |
| Amplicon sequencing | AmpliSeq for VR���˲�Ʊ Panels | AmpliSeq UD Indexes for VR���˲�Ʊ (for ≤ 24 samples) AmpliSeq UD Indexes for VR���˲�Ʊ (for > 24 samples) |
| Targeted DNA enrichment | VR���˲�Ʊ DNA Prep with Enrichment | IDT for VR���˲�Ʊ Nextera UD Indexes |
Frequently Purchased Together
Ask an expert how you can get the most with VR���˲�Ʊ Library Prep
We'll walk through your needs and make recommendations.
Should I Use Unique Dual Indexes or Unique Molecular Identifiers?
Unique dual indexes are useful when multiplexing to mitigate sample misassignment due to index hopping. UDIs can be especially effective when using instruments with patterned flow cells, such as the NovaSeq 6000 system.
Unique molecular identifiers have been shown to reduce the rate of false-positive variant calls and increase sensitivity of variant detection. They are useful for removing PCR duplicates in DNA and cDNA and for detection of low-frequency variants such as highly-multiplexed CNV detection. Learn more about unique molecular identifiers.
Single indexing vs dual indexing
Generally speaking, dual indexing is the preferred route. Dual index sequencing does require extra cycles; however, the reagent kits supply enough to perform the extra cycles. You might still want to perform single indexing if:
- The instrument does not support dual sequencing (HiSeq 1000/1500 systems)
- The reads need to be a few bases longer
- Your schedule does not permit an additional 1-2 hours required to run the second index

NovaSeq 6000 System
The NovaSeq 6000 System unites the latest high-performance imaging with VR���˲�Ʊ patterned flow cell technology. The updated flow cell design reduces spacing between nanowells, significantly increasing cluster density and data output.
Resources for Dual Index Sequencing
VR���˲�Ʊ Adapter Sequences
This document contains the oligonucleotide sequences of VR���˲�Ʊ adapters used in several VR���˲�Ʊ library prep kits.
Indexed Sequencing Guide
Learn about single-indexed and dual-indexed sequencing on VR���˲�Ʊ instruments.
VR���˲�Ʊ Adapters Pooling Guide
This document provides recommendations for optimizing color balance across all VR���˲�Ʊ systems when pooling indexed libraries.
VR���˲�Ʊ Adapters Support
Find helpful resources and support information for IDT for VR���˲�Ʊ unique dual indexes.